Research in Biomedical Image Analysis at IDSIA
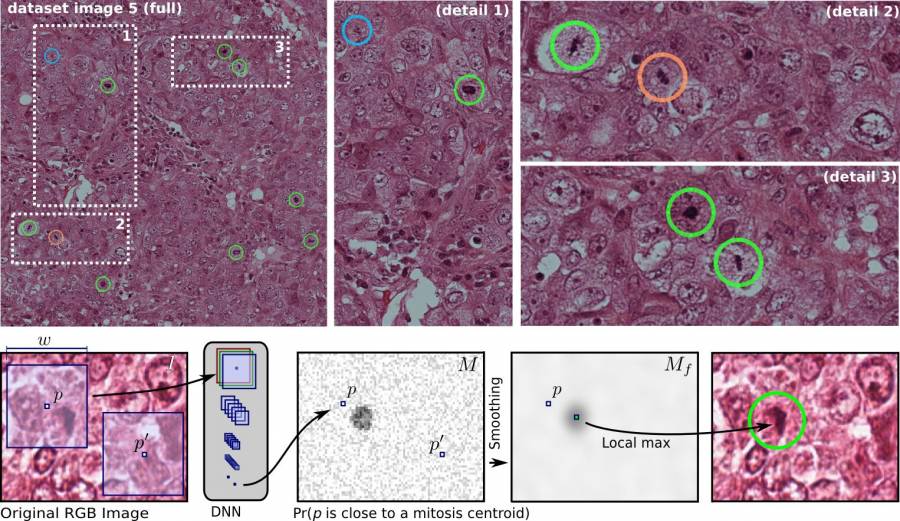
Mitosis Detection in Breast Cancer Histology Images using Deep Neural Networks
Joint work with Dr. Dan Ciresan.
- Our research was featured in the widely popular two minute papers youtube channel!
- The Medical Image Analysis paper describing the MICCAI 2013 Grand Challenge “Assessment of Mitosis Detection Algorithms” is available here. With Dan Ciresan, we won that challenge with a significant advantage over the closest competitor; we also won the ICPR 2012 challenge on mitosis detection in breast cancer histology images. The paper describing the approach was published at MICCAI 2013.
Related publications
- A. Giusti, C. Caccia, D. Ciresan, J. Schmidhuber, L. Gambardella: “A Comparison of Algorithms and Humans for Mitosis Detection”. Proc. of International Symposium on Biomedical Imaging (ISBI) 2014. oral presentation.
- D. Ciresan, A. Giusti, L. Gambardella, J. Schmidhuber: “Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks”. Proc. of International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2013. Describing our winning entry to ICPR 2012 Mitosis Detection competition. pubmed
- M. Veta et al.: “Assessment of algorithms for mitosis detection in breast cancer histopathology images”. Medical Image Analysis. (preprint, pubmed, arxiv).
- M. Veta, P. J. van Diest, D. Ciresan, A. Giusti, J. Pluim: “Prognostic value of automatic mitosis detection in breast cancer histopathology images”. Proc. of International Symposium on Biomedical Imaging (ISBI) 2015. (abstract).
@inproceedings{Giusti2014comparison,
author = {Alessandro Giusti and
Dan Claudiu Ciresan and
Claudio Caccia and
J{\"u}rgen Schmidhuber and
Luca Maria Gambardella},
title = {A Comparison of Algorithms and Humans for Mitosis Detection},
booktitle = {International Symposium on Biomedical Imaging (ISBI)},
year = {2014}
}
@inproceedings{Ciresan2013mitosis,
author = {Dan Claudiu Ciresan and
Alessandro Giusti and
Luca Maria Gambardella and
J{\"u}rgen Schmidhuber},
title = {Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks},
booktitle = {MICCAI},
year = {2013},
volume = {2},
pages = {411-418}
}
@article{Veta2015assessment,
title={Assessment of algorithms for mitosis detection in breast cancer histopathology images},
author={Veta, Mitko and van Diest, Paul J and Willems, Stefan M and Wang, Haibo and Madabhushi, Anant and Cruz-Roa, Angel and Gonzalez, Fabio and Larsen, Anders BL and Vestergaard, Jacob S and Dahl, Anders B and others},
journal={Medical image analysis},
volume={20},
number={1},
pages={237--248},
year={2015},
publisher={Elsevier}
}
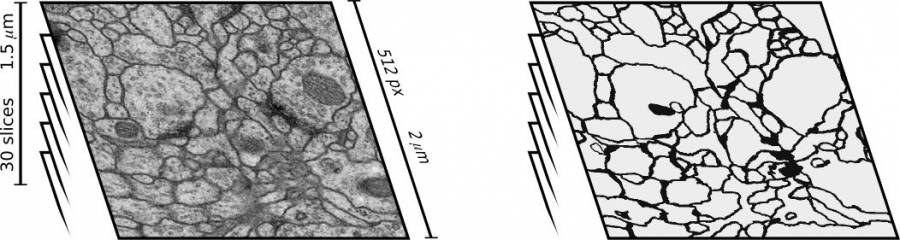
Segmentation of Neural Structures from Electron Microscopy Stacks using Deep Neural Networks
Joint work with Dr. Dan Ciresan and other colleagues at MIT, Janelia Farm, and ETH.
- We won (by a large margin) the ISBI 2012 challenge on segmentation of neuronal structures in electron microscopy stacks. Our entry outperforms all the 16 competing methods on each of the three considered metrics. In addition, we are the only entry outperforming humans in the pixel error metric. We published our approach at NIPS 2012.
Related publications
- D. Ciresan, A. Giusti, L. Gambardella, J. Schmidthuber: “Deep Neural Networks Segment Neuronal Membranes in Electron Microscopy Images”. Proc. of International Conference on Neural Information Processing Systems (NIPS) 2012 (acceptance rate ~25%).
- T. Parag, D. Ciresan, A. Giusti: “Efficient Classifier Training to Reduce False Merges in Electron Microscopy Segmentation”. Proc. of International Conference on Computer Vision (ICCV) 2015
- I. Arganda-Carreras et al.: “Crowdsourcing the creation of image segmentation algorithms for connectomics”. Frontiers in Neuroanatomy 2015. (DOI)
- J. Funke, et al.: “Candidate Sampling for Neuron Reconstruction from Anisotropic Electron Microscopy Volumes”. Proc. of International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2014
- D. Ciresan, A. Giusti, L. Gambardella, J. Schmidthuber: “Neural Networks for Segmenting Neuronal Structures in EM Stacks”. Describing our winning entry (in all three metrics) for ISBI 2012 competition “Segmentation of neuronal structures in EM stacks”, Workshop at International Symposium on Biomedical Imaging (ISBI) 2012.
@inproceedings{Parag:2015,
title={Efficient Classifier Training to Reduce False Merges in Electron Microscopy Segmentation},
author={Parag, Toufiq and Cire{\c{s}}an, Dan C and Giusti, Alessandro},
booktitle={ICCV},
year={2015}
}
@inproceedings{Funke:2014,
title={Candidate Sampling for Neuron Reconstruction from Anisotropic Electron Microscopy Volumes},
author={Funke, Jan and Martel, Julien NP and Gerhard, Stephan and Andres, Bjoern and Cire{\c{s}}an, Dan C and Giusti, Alessandro and Gambardella, Luca M and Schmidhuber, J{\"u}rgen and Pfister, Hanspeter and Cardona, Albert and others},
booktitle={Medical Image Computing and Computer-Assisted Intervention--MICCAI 2014},
pages={17--24},
year={2014},
publisher={Springer}
}
@inproceedings{Ciresan:2012f,
author = {Dan Claudiu Ciresan and
Alessandro Giusti and
Luca Maria Gambardella and
J{\"u}rgen Schmidhuber},
title = {Deep Neural Networks Segment Neuronal Membranes in Electron Microscopy Images},
booktitle = {NIPS},
year = {2012},
pages = {2852--2860}
}
@inproceedings{Ciresan:2012d,
author = {Dan Claudiu Ciresan and
Alessandro Giusti and
Luca Maria Gambardella and
J{\"u}rgen Schmidhuber},
title = {Neural Networks for Segmenting Neuronal Structures in EM Stacks},
booktitle = {Segmentation of Neuronal Structures in EM Stacks Workshop at International Symposium on Biomedical Imaging},
year = {2012}
}
Image Analysis (and Statistical Modeling) for Human Embryo Grading
Joint work with [Giorgio Corani(http://www.idsia.ch/~giorgio) and the International Institute of Reproductive Medicine in Lugano, Switzlerland.
We worked on the segmentation of zygotes (1 cell, shortly after fertilization), and embryos up to 4 cells, from Hoffmann Modulation Contrast (HMC) focus stacks. It is a mostly unexplored and challenging imaging modality from the point of view of automated processing; focus stacks are also an unusual and interesting type of data. Giorgio has focused on the statistical modeling of the embryo selection problem.
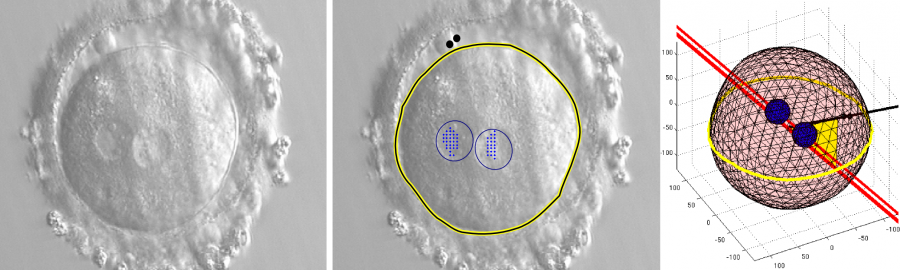
Zygote Segmentation and 3D Reconstruction
Related publications
- A. Giusti, G. Corani, L. Gambardella, C. Magli, L. Gianaroli: “3D Localization of Pronuclei of Human Zygotes using Texture Features from Multiple Focal Planes”. Proc. of International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2010.
- A. Giusti, G. Corani, L. Gambardella, C. Magli, L. Gianaroli: “Symultaneous Focusing and Contouring of Human Zygotes for In Vitro Fertilization”. BIOSIGNALS 2010. Oral presentation (acceptance rate ~39%).
- A. Giusti et al.: “Lighting-Aware Segmentation of Microscopy Images for In Vitro Fertilization”. Proc. of International Symposium on Visual Computing (ISVC) 2009. Oral presentation (acceptance rate ~30%).
- A. Giusti et al.: “Segmentation of Human Zygotes in Hoffman Modulation Contrast Images”. Proc. of Medical Image Understanding and Analysis (MIUA) 2009. Best Poster Award.
@incollection{giusti20103d,
title={3D localization of pronuclei of human zygotes using textures from multiple focal planes},
author={Giusti, Alessandro and Corani, Giorgio and Gambardella, L and Magli, Cristina and Gianaroli, Luca},
booktitle={Medical Image Computing and Computer-Assisted Intervention--MICCAI 2010},
pages={488--495},
year={2010},
publisher={Springer}
}
@inproceedings{giusti2010blastomere,
title={Blastomere segmentation and 3d morphology measurements of early embryos from hoffman modulation contrast image stacks},
author={Giusti, Alessandro and Corani, Giorgio and Gambardella, Luca and Magli, Cristina and Gianaroli, Luca},
booktitle={Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on},
pages={1261--1264},
year={2010},
organization={IEEE}
}
@incollection{giusti2009lighting,
title={Lighting-Aware Segmentation of Microscopy Images for In Vitro Fertilization},
author={Giusti, Alessandro and Corani, Giorgio and Gambardella, Luca Maria and Magli, Cristina and Gianaroli, Luca},
booktitle={Advances in Visual Computing},
pages={576--585},
year={2009},
publisher={Springer}
}
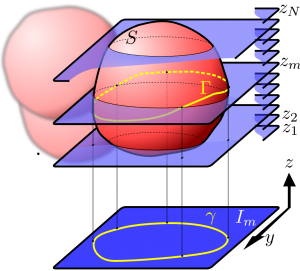
4-Cell Embryo Segmentation and 3D Reconstruction
Monitoring of embryos is an important activity during In Vitro Fertilization (IVF) procedures. A critical stage is that of the early embryo, which is formed by a small number of cells (blastomeres): the morphology of such cells is considered to be a powerful indicator of the embryo viability. We deal with the challenging problem of automatically segmenting the different blastomeres in the early embryo while simultaneously determining their depth, by processing a Z-stack of images acquired by means of an Hoffman Modulation Contrast (HMC) microscope. We discuss experimental results on 53 embryo image stacks, and elaborate on the advantages and limitations of our approach, also briefly describing it is being integrated in the workflow of an IVF laboratory.
Practical application of these techniques prompted the implementation of a tool for the Visualization of Markers in Microscopy Z-Stacks.
Related Publications
- A. Giusti, G. Corani, L. Gambardella, C. Magli, L. Gianaroli: “Blastomere Segmentation and 3D Morphology Measurements of Early Embryos from Hoffman Modulation Contrast Image Stacks”. Proc of IEEE International Symposium on Biomedical Imaging (ISBI) 2010.
- A. Giusti, G. Corani, L. Gambardella, C. Magli, L. Gianaroli: “Segmenting Embryo Blastomeres in 3D from Hoffman Modulation Contrast Image Stacks”. Proc. of Medical Image Understanding and Analysis (MIUA) 2010.
- A. Giusti, C. Magli, G. Corani, L. Gambardella, L. Gianaroli: “Observing the 3-Dimensional Morphology of 4-Cell Embryos using Computer Analysis of Image Z-Stacks”. Human Reproduction (Supplement: Abstract book) vol.26 2011. Oral presentation @ European Society on Human Reproduction and Embryology (ESHRE) Annual Meeting 2011 (acceptance rate < 18%).
@inproceedings{giusti2010blastomere,
title={Blastomere segmentation and 3d morphology measurements of early embryos from hoffman modulation contrast image stacks},
author={Giusti, Alessandro and Corani, Giorgio and Gambardella, Luca and Magli, Cristina and Gianaroli, Luca},
booktitle={Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on},
pages={1261--1264},
year={2010},
organization={IEEE}
}
@inproceedings{giusti2011observing,
title={Observing the 3-dimensional morphology of 4-cell embryos using computer analysis of image z-stacks},
author={Giusti, A and Magli, MC and Corani, G and Paviglianiti, B and Gianaroli, L and Gambardella, L},
booktitle={Human Reproduction},
volume={26},
pages={I62--I63},
year={2011}
}
Statistics
Publications (more details at Giorgio Corani’s website:
- L. Gianaroli, C. Magli, L. Gambardella, A. Giusti, C. Grugnetti, G. Corani: “Objective Way to Support Embryo Transfer: a Probabilistic Decision”. Human Reproduction, 2013 (impact factor 4.47).
- G. Corani, A. Giusti, C. Magli, et al: “A prognostic Model for Multiple-embryo Transfers”. Human Reproduction (Supplement: Abstract book). Presentation @ European Society on Human Reproduction and Embryology (ESHRE) Annual Meeting 2012.
- G. Corani, C. Magli, A. Giusti, L. Gianaroli, L. Gambardella: “A Bayesian Network Model for Predicting the Outcome of In Vitro Fertilization”. Proc. of European Workshop on Probabilistic Graphical Models (PGM), 2012 (acceptance rate ~45%).
Novel Statistical Methods for Analyzing Images of Fibre-like Structures
Joint work with Paola Rancoita.
- A. Giusti, P. Rancoita: “Quantifying Challenging Images of Fiber-Like Structures” Proc. of IEEE International Conference on Image Processing (ICIP) 2013
- P. Rancoita, A. Giusti: “On Estimating the Intensity of Stationary Fibre Processes from Real Digital Images” Oral presentation at European Congress of Stereology and Image Analysis, 2013
- P.M.V. Rancoita, A. Giusti, A. Micheletti: “Intensity estimation of stationary fibre processes from digital images with a learned detector”. Image Analysis and Stereology 30(3):167-178, 2011.
- P.M.V. Rancoita, A. Giusti, A. Micheletti: “A General Technique for the Estimation of the Intensity of Stationary Fibre Processes from Digital Images using a Learned Detector”. Proc. of Shape and Size in Medicine, Biotechnology, Materials Science and Social Sciences, abstract, 2011.
Segmentation of micro-CT volumes
Joint work with Dr. Anderson Meneses
- A. Meneses, A. Giusti, A. de Almeida, L. Nogueira, D. Braz, R. Barroso, C. de Almeida: “Automated Segmentation of Synchrotron Radiation Micro-Computed Tomography Biomedical Images using Graph Cuts and Neural Networks”. Nuclear Instruments and Methods in Physics Research, 2011.
